DNA Data Storage Overview
DNA, the blueprint of life, is not only responsible for encoding genetic information but also holds incredible potential as a data storage medium to store archival data in the long term. Find out more about DNA data storage and the research we do at Imperial in the two videos below which feature our research and the remainder of the content. Do not forget to read our DNA storage FAQ and read about our work in a recent article in Chemistry World to which we contributed!
Read our DNA Storage FAQOur Research Focus
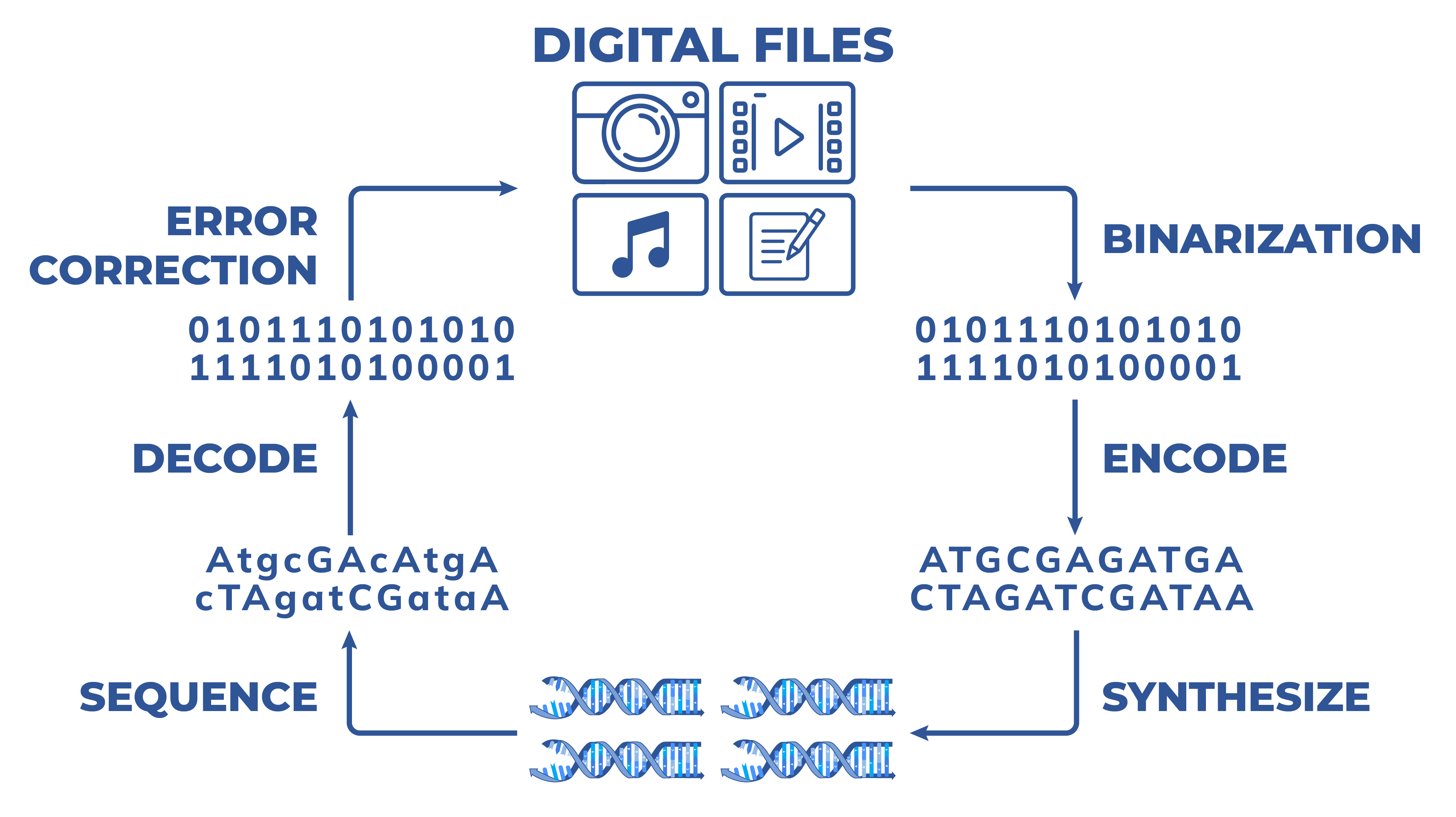
DNA data storage starts with translating any type of binary files (documents, movies, music, databases) to a binary representation, encoding the binary representation to sequences while incorporating error correction, and then synthesising the sequence representation to physical DNA molecules. The resulting molecules may be stored for years if not decades after which they are sequenced to recover the data. The resulting sequencing data is aligned, decoded and potential errors are fixed based on incorporated error correction codes to finally obtain the digital files encoded.
Our research focuses on all stages of the DNA data storage cycle:
– Encoding, decoding and error correction for DNA data storage
– Optimising synthesis processes for DNA data storage
– Physical storage of DNA molecules for data storage
– Sequencing for DNA storage based on Machine Learning/AI methods
Making DNA Data Storage a Reality
Our ultimate goal to make it a staple of modern data centres to store archival data in the very long term.



DNA Data Storage
DNA Data Storage, the process of storing arbitrary binary data in DNA sequences is actively researched by us and the community. Its properties, primarily durability and compact form factor, make it an ideal storage media for archiving data for decades. Watch the video here to see what makes DNA an ideal storage media and how it all works.
Processing Data in DNA
Once data is stored in DNA, biomolecular processes can be used to process it on an unprecedented scale. More precisely, combinatorial problems (e.g., databases joins, travelling salesperson problems and others) can be solved in DNA very fast thanks to the unprecedented level of parallelism and also very energy efficient comapred to traditional computing. Watch the video here to learn more.
Modelling DNA Storage as a Constrained Channel
Key to storing binary data in synthetic in DNA is the translation between the binary representation of digital data to the quaternary domain of DNA. This translation must adhere to constraints imposed by the synthesis and sequencing processes used to write and read respectively. A technological advance in either process changes the constraints and renders current encoding schemes obsolete. In this line of work we present a recipe for taking constraints and producing an appropriate encoding scheme. Such a mechanism allows moving the encoding in lockstep with the technological advances in the underlying processes. We further show a method to understand trade-offs in constraints for a given overhead of bits needed to meet such constraints
Storing Data in DNA Nanostructures
Synthetic DNA has garnered attention as a storage medium due to its high density and durability. DNA has crucial properties that make it pertinent for archival storage. However, synthetic DNA is currently prohibitively expensive and so alternative forms of DNA storage need to be explored. Given the prohibitive cost of synthetic DNA, our vision is to use DNA nanostructures with molecular bumps to store data—in a similar manner to a compact disc—and to use microscopy, followed by a decoding pipeline based on computer vision and machine learning, to reconstruct the original data. The following picture illustrates our approach.
Publications
Selected publications resulting from our research
2022
Generating Synthetic Data for DNA Origami-based Information Storage Systems Conference
2022.
2021
DNA archival storage, a bottom up approach Conference
ACM Workshop on Hot Topics in storage and File Systems, 2021.
DNA4DNA: Preserving Culturally Significant Digital Data with Synthetic DNA Journal Article
In: 17th International Conference on Digital Preservation (iPRES 2021), 2021.
Digital Preservation with Synthetic DNA Journal Article
In: 37eme Conference sur la Gestion de Donnees – Principes, Technologies et Applications (BDA 2021), 2021.
Nanopore Sequencing Simulator or DNA Data Storage Journal Article
In: Visual Communications and Image Processing (VCIP 2021), 2021.
2019
OligoArchive: Using DNA in the DBMS Storage Hierarchy. Proceedings Article
In: Conference on Innovative Data Systems Research (CIDR '19) , 2019.
Survey of information encoding techniques for dna Journal Article
In: arXiv preprint arXiv:1906.11062, 2019.
Meet the Team
Join us! We have the following student, PhD and PostDoc opportunities available…
Partners & Collaborators













